

|
latest version v1.9 - last update 10 Apr 2010 |
|
This class finds and evaluates the best path through a Hidden Markov Model (HMM) for a given observation sequence  .
More...
.
More...
#include <ltiHmmViterbiPathSearch.h>
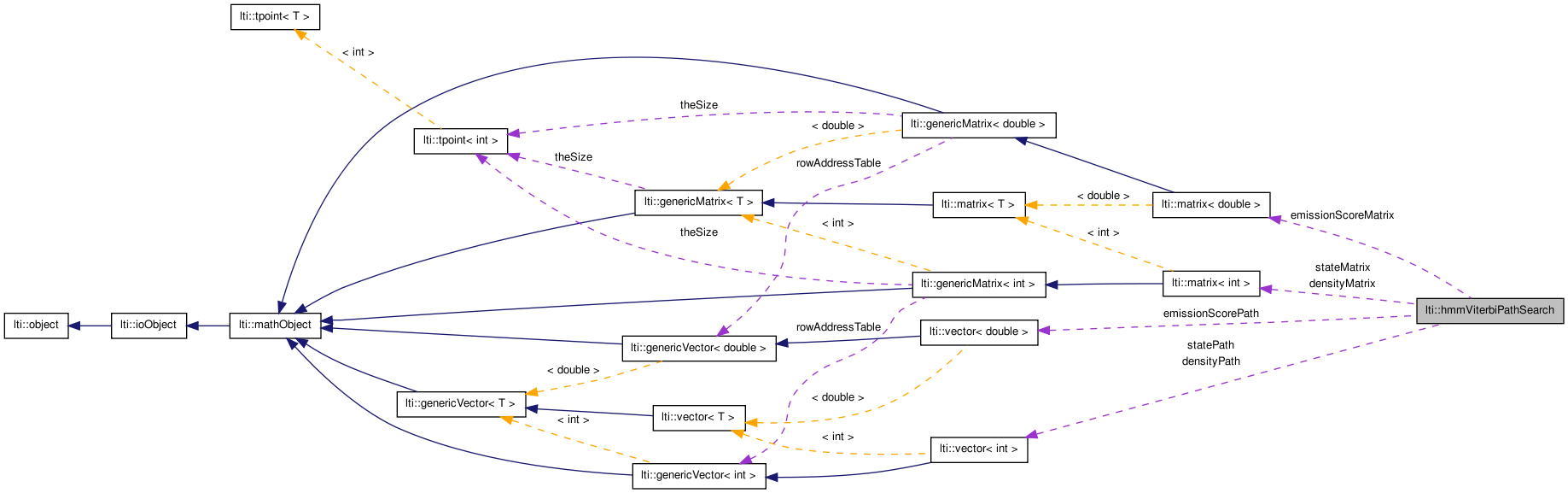
Public Types | |
| typedef sequence< dvector > | observationSequence |
Public Member Functions | |
| hmmViterbiPathSearch () | |
| hmmViterbiPathSearch (const hmmViterbiPathSearch &other) | |
| virtual | ~hmmViterbiPathSearch () |
| virtual const char * | getTypeName () const |
| hmmViterbiPathSearch & | copy (const hmmViterbiPathSearch &other) |
| double | apply (const hiddenMarkovModel &model, const observationSequence &srcData) |
| double | evaluatePath (const hiddenMarkovModel &model, const observationSequence &srcData, const ivector &path) |
| double | evaluatePath (const hiddenMarkovModel &model, const observationSequence &srcData, const ivector &path, const ivector &densityPath) |
| double | minimumPathScore (const hiddenMarkovModel &model, int pathLength) const |
| double | minimumPathScore (const hiddenMarkovModel &model, const ivector &stateSelection, const ivector &densitySelection=ivector()) const |
State selection logging | |
These methods can be used to enable/disable logging and access the selection of states Default is "disabled", for efficiency reasons. Enable only if necessary. | |
| void | logStateSelection (bool enable=true) |
| const matrix< int > & | getBackpointerMatrix () const |
| const vector< int > & | getStatePath () |
Density selection logging | |
These methods can be used to enable/disable logging and access the selection of densities during a call of apply(). Density selection stands for the centre | |
| void | logDensitySelection (bool enable=true) |
| const matrix< int > & | getDensitySelectionMatrix () const |
| const vector< int > & | getDensitySelectionPath () |
Emission score logging | |
These methods can be used to enable/disable logging and access the emission scores Default is "disabled", for efficiency reasons. Enable only if necessary. | |
| void | logEmissionScore (bool enable=true) |
| const matrix< double > & | getEmissionScoreMatrix () const |
| const vector< double > & | getEmissionScorePath () |
Routines for online classification | |
| double | initialStep (const hiddenMarkovModel &model, const dvector &frame, dvector &result) const |
| double | subsequentStep (const hiddenMarkovModel &model, const dvector &frame, dvector &result) const |
This class finds and evaluates the best path through a Hidden Markov Model (HMM) for a given observation sequence  .
.
The best path is found with the Viterbi algorithm. The algorithm is taken from "Fundamentals of speech recognition" (L. Rabiner and B.-H. Juang, Prentice Hall, 1993) and can also be found in "Erkennung kontinuierlicher Gebärdensprache mit Ganzwortmodellen" (H. C. Hienz, PhD thesis, Chair of Technical Computer Science, 2000). The notation in this documentation is taken from the latter literature.
Increased computational efficiency and precision are achieved by using scores with 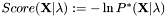 instead of probabilities. Note that the lowest score corresponds to the highest probability and thus represents the best recognition result.
instead of probabilities. Note that the lowest score corresponds to the highest probability and thus represents the best recognition result.
Viterbi algorithm:
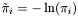
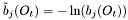
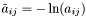
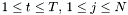
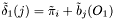
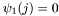
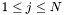
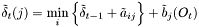
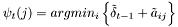
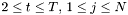
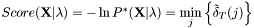
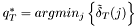
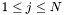
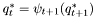
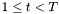
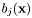 is approximated by the centre of the distribution, which provides the best estimation of the observation, as follows:
is approximated by the centre of the distribution, which provides the best estimation of the observation, as follows: 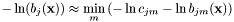 .
. 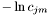 denotes the weight scores of the mixture densities
denotes the weight scores of the mixture densities 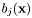 . All components
. All components 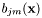 of
of 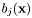 are modelled by either laplace or gaussian distributions.
are modelled by either laplace or gaussian distributions. 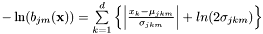
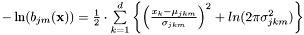
An alternative notation for the score of the best path 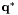 is
is 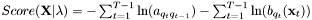 .
.
Note: Each state sequence 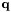 always starts in the first state
always starts in the first state 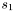 and ends in the last state
and ends in the last state 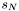 , whereas
, whereas 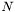 denotes the number of states
denotes the number of states 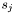 . Thus, the termination of the Viterbi algorithm is implemented as
. Thus, the termination of the Viterbi algorithm is implemented as 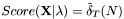 .
.
Note: The preprocessing values 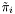 ,
, 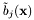 and
and 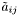 are usually estimated over a given data set by an automatic training algorithm (see hmmTrainer).
are usually estimated over a given data set by an automatic training algorithm (see hmmTrainer).
type of an observation sequence
| lti::hmmViterbiPathSearch::hmmViterbiPathSearch | ( | ) |
default constructor
| lti::hmmViterbiPathSearch::hmmViterbiPathSearch | ( | const hmmViterbiPathSearch & | other | ) |
copy constructor
| other | the object to be copied |
| virtual lti::hmmViterbiPathSearch::~hmmViterbiPathSearch | ( | ) | [virtual] |
destructor
| double lti::hmmViterbiPathSearch::apply | ( | const hiddenMarkovModel & | model, | |
| const observationSequence & | srcData | |||
| ) |
| hmmViterbiPathSearch& lti::hmmViterbiPathSearch::copy | ( | const hmmViterbiPathSearch & | other | ) |
| double lti::hmmViterbiPathSearch::evaluatePath | ( | const hiddenMarkovModel & | model, | |
| const observationSequence & | srcData, | |||
| const ivector & | path, | |||
| const ivector & | densityPath | |||
| ) |
Calculates score along a given path for a given (observed) data sequence, and additionally selects the specified density centers of the mixture densities in the visited path.
| model | the hidden markov model | |
| srcData | contains the data sequence. | |
| path | defines the applicable state sequence. | |
| densityPath | contains the applicable mixture density center selection. |
| double lti::hmmViterbiPathSearch::evaluatePath | ( | const hiddenMarkovModel & | model, | |
| const observationSequence & | srcData, | |||
| const ivector & | path | |||
| ) |
| const matrix<int>& lti::hmmViterbiPathSearch::getBackpointerMatrix | ( | ) | const |
| const matrix<int>& lti::hmmViterbiPathSearch::getDensitySelectionMatrix | ( | ) | const |
| const vector<int>& lti::hmmViterbiPathSearch::getDensitySelectionPath | ( | ) |
| const matrix<double>& lti::hmmViterbiPathSearch::getEmissionScoreMatrix | ( | ) | const |
| const vector<double>& lti::hmmViterbiPathSearch::getEmissionScorePath | ( | ) |
| const vector<int>& lti::hmmViterbiPathSearch::getStatePath | ( | ) |
| virtual const char* lti::hmmViterbiPathSearch::getTypeName | ( | ) | const [virtual] |
returns the name of this type ("hmmViterbiPathSearch")
| double lti::hmmViterbiPathSearch::initialStep | ( | const hiddenMarkovModel & | model, | |
| const dvector & | frame, | |||
| dvector & | result | |||
| ) | const |
perform initial transition and add emission score, i.e.
initialisation of the Viterbi algorithm
| void lti::hmmViterbiPathSearch::logDensitySelection | ( | bool | enable = true |
) |
| void lti::hmmViterbiPathSearch::logEmissionScore | ( | bool | enable = true |
) |
Turns logging of emission score matrix on and off.
This enables to analyse the emission scores generated for each step and state
By default, this is turned off for efficiency reasons (runtime and memory).
| void lti::hmmViterbiPathSearch::logStateSelection | ( | bool | enable = true |
) |
Turns logging of state selection on and off.
This enables to review the state matrix and the path leading to the minimal score.
By default, this is turned off for efficiency reasons (memory).
| double lti::hmmViterbiPathSearch::minimumPathScore | ( | const hiddenMarkovModel & | model, | |
| const ivector & | stateSelection, | |||
| const ivector & | densitySelection = ivector() | |||
| ) | const |
find out lowest possible score of the given model for a given path.
| model | the hidden markov model | |
| stateSelection | the state selection along the given path | |
| densitySelection | the density selection along the given path. if this is empty, the best densities are chosen. |
| double lti::hmmViterbiPathSearch::minimumPathScore | ( | const hiddenMarkovModel & | model, | |
| int | pathLength | |||
| ) | const |
| double lti::hmmViterbiPathSearch::subsequentStep | ( | const hiddenMarkovModel & | model, | |
| const dvector & | frame, | |||
| dvector & | result | |||
| ) | const |
perform subsequent transition and add emission score, i.e.
one iteration of the Viterbi algorithm recursion