

|
latest version v1.9 - last update 10 Apr 2010 |
|
the parameters for the class MSTClustering More...
#include <ltiMSTClustering.h>
Public Types | |
| enum | eProbabilityFromDistanceTypes |
Public Member Functions | |
| parameters () | |
| parameters (const parameters &other) | |
| ~parameters () | |
| const char * | getTypeName () const |
| parameters & | copy (const parameters &other) |
| parameters & | operator= (const parameters &other) |
| virtual classifier::parameters * | clone () const |
| bool | write (ioHandler &handler, const bool complete=true) const |
| bool | read (ioHandler &handler, const bool complete=true) |
Public Attributes | |
| float | devFac |
| int | nbMaxSteps |
| double | variance |
| eProbabilityFromDistanceTypes | probabilityFromDistanceMode |
the parameters for the class MSTClustering
| enum lti::MSTClustering::parameters::eProbabilityFromDistanceTypes |
The different methods that can be used to compute a probability from a distance.
| lti::MSTClustering< U >::parameters::parameters | ( | void | ) | [inline] |
default constructor
Reimplemented from lti::clustering::parameters.
References lti::MSTClustering< U >::parameters::devFac, lti::MSTClustering< U >::parameters::nbMaxSteps, lti::MSTClustering< U >::parameters::probabilityFromDistanceMode, and lti::MSTClustering< U >::parameters::variance.
Referenced by lti::MSTClustering< U >::parameters::clone().
| lti::MSTClustering< U >::parameters::parameters | ( | const parameters & | other | ) | [inline] |
copy constructor
| other | the parameters object to be copied |
Reimplemented from lti::clustering::parameters.
References lti::MSTClustering< U >::parameters::copy().
| lti::MSTClustering< U >::parameters::~parameters | ( | ) | [inline, virtual] |
destructor
Reimplemented from lti::clustering::parameters.
| virtual classifier::parameters* lti::MSTClustering< U >::parameters::clone | ( | ) | const [inline, virtual] |
returns a pointer to a clone of the parameters
Reimplemented from lti::clustering::parameters.
References lti::MSTClustering< U >::parameters::parameters().
| parameters& lti::MSTClustering< U >::parameters::copy | ( | const parameters & | other | ) | [inline] |
copy the contents of a parameters object
| other | the parameters object to be copied |
Reimplemented from lti::clustering::parameters.
References lti::clustering::parameters::copy(), lti::MSTClustering< U >::parameters::devFac, lti::MSTClustering< U >::parameters::nbMaxSteps, lti::MSTClustering< U >::parameters::probabilityFromDistanceMode, and lti::MSTClustering< U >::parameters::variance.
Referenced by lti::MSTClustering< U >::parameters::operator=(), and lti::MSTClustering< U >::parameters::parameters().
| const char* lti::MSTClustering< U >::parameters::getTypeName | ( | void | ) | const [inline, virtual] |
returns name of this type
Reimplemented from lti::clustering::parameters.
| parameters& lti::MSTClustering< U >::parameters::operator= | ( | const parameters & | other | ) | [inline] |
copy the contents of a parameters object
| other | the parameters object to be copied |
Reimplemented from lti::unsupervisedClassifier::parameters.
References lti::MSTClustering< U >::parameters::copy().
| bool lti::MSTClustering< U >::parameters::read | ( | ioHandler & | handler, | |
| const bool | complete = true | |||
| ) | [inline, virtual] |
read the parameters from the given ioHandler
| handler | the ioHandler to be used | |
| complete | if true (the default) the enclosing begin/end will be also written, otherwise only the data block will be written. |
Reimplemented from lti::clustering::parameters.
References lti::MSTClustering< U >::parameters::devFac, lti::MSTClustering< U >::parameters::nbMaxSteps, lti::MSTClustering< U >::parameters::probabilityFromDistanceMode, lti::ioHandler::readBegin(), lti::ioHandler::readEnd(), and lti::MSTClustering< U >::parameters::variance.
| bool lti::MSTClustering< U >::parameters::write | ( | ioHandler & | handler, | |
| const bool | complete = true | |||
| ) | const [inline, virtual] |
write the parameters in the given ioHandler
| handler | the ioHandler to be used | |
| complete | if true (the default) the enclosing begin/end will be also written, otherwise only the data block will be written. |
Reimplemented from lti::clustering::parameters.
References lti::MSTClustering< U >::parameters::devFac, lti::MSTClustering< U >::parameters::nbMaxSteps, lti::MSTClustering< U >::parameters::probabilityFromDistanceMode, lti::MSTClustering< U >::parameters::variance, lti::ioHandler::writeBegin(), and lti::ioHandler::writeEnd().
| float lti::MSTClustering< U >::parameters::devFac |
Factor that is used to decide if an edge is inconsistent.
First all edges are searched that lie at most nbMaxSteps away from the current node. Then m is the mean of the weights of all these edges and sigma is the standard deviation of these edges. An Edge is considered to be in consistent if is weight is bigger than mean+devFac*sigma. Default is 2.0
Referenced by lti::MSTClustering< U >::parameters::copy(), lti::MSTClustering< U >::parameters::parameters(), lti::MSTClustering< U >::parameters::read(), and lti::MSTClustering< U >::parameters::write().
| int lti::MSTClustering< U >::parameters::nbMaxSteps |
The maximum number of edges that can be between the starting point and the last edge that is considered when computed inconsistent edges.
Default is 4.
Referenced by lti::MSTClustering< U >::parameters::copy(), lti::MSTClustering< U >::parameters::parameters(), lti::MSTClustering< U >::parameters::read(), and lti::MSTClustering< U >::parameters::write().
| eProbabilityFromDistanceTypes lti::MSTClustering< U >::parameters::probabilityFromDistanceMode |
The method used to compute a probabilty from the distance between the feature that is classified and the clusters.
Valid valued are: InterClusterDistanceDependend and InterClusterDistanceIndependend. See parameter variance
Referenced by lti::MSTClustering< U >::parameters::copy(), lti::MSTClustering< U >::parameters::parameters(), lti::MSTClustering< U >::parameters::read(), and lti::MSTClustering< U >::parameters::write().
| double lti::MSTClustering< U >::parameters::variance |
Variance used to compute a possibility from the distances for classification.
While classification, the membership of feature to all clusters is computed. This done by the following: Say d is the minimal distance between the feature that is classified and one of the clusters. Say also w is the the minimal distance between the current cluster the nearest neighboring cluster, then the membership is defined either 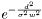 or
or 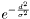 depending on the value of probabilityFromDistanceMode Default is 0.33333.
depending on the value of probabilityFromDistanceMode Default is 0.33333.
Referenced by lti::MSTClustering< U >::parameters::copy(), lti::MSTClustering< U >::parameters::parameters(), lti::MSTClustering< U >::parameters::read(), and lti::MSTClustering< U >::parameters::write().